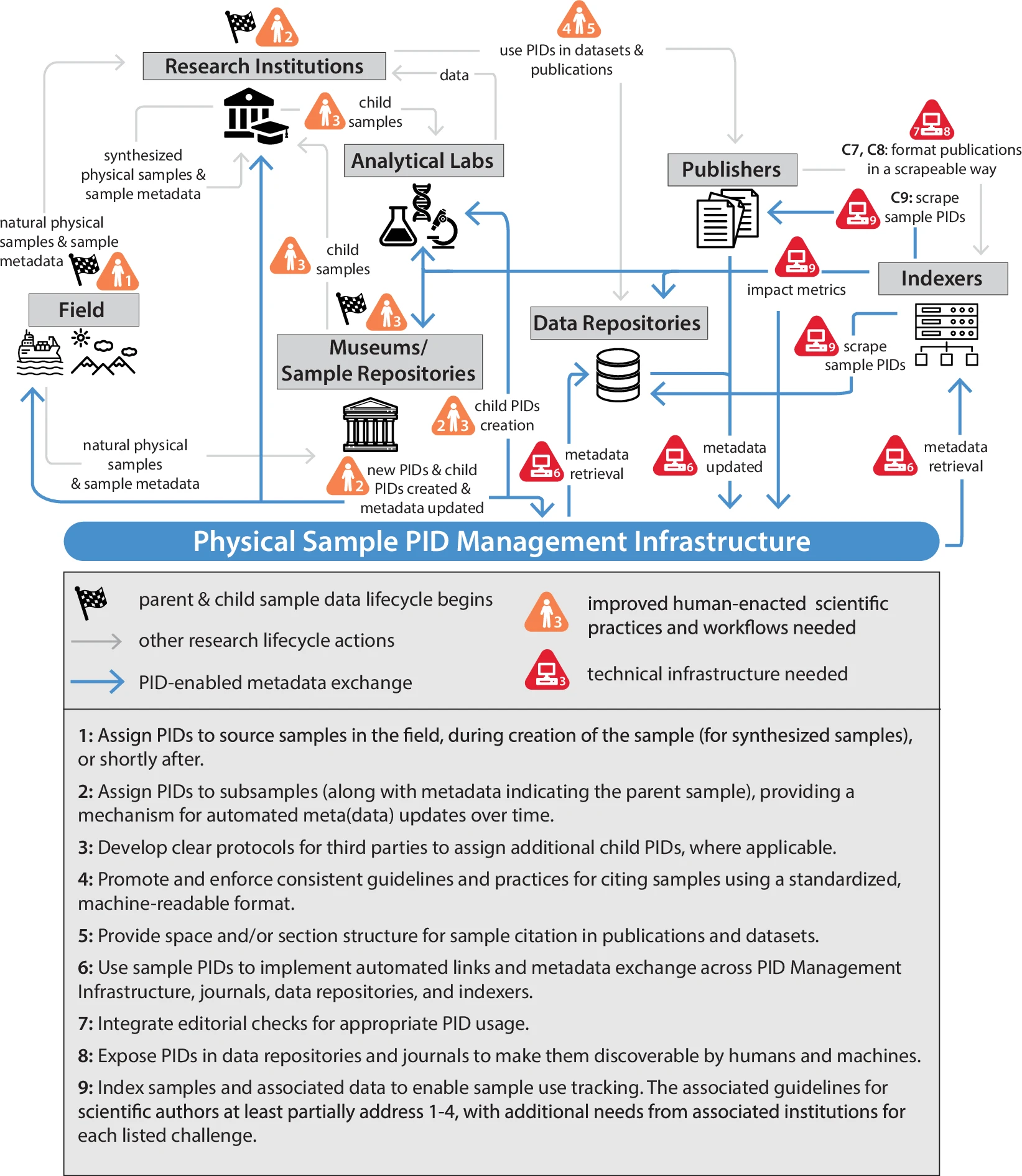
Workflow diagram showing the integration of persistent identifiers in sample tracking and research attribution. Source: Damerow et al., Scientific Data (2025)
Imagine a world where every rock sample collected from a remote mountain, every soil core extracted from a wetland, and every water sample drawn from a river could tell its complete story—not just where it came from, but every analysis performed, every dataset created, and every scientific discovery it enabled. This vision is closer to reality thanks to groundbreaking new guidelines from the Earth Science Information Partners (ESIP) Physical Samples Curation Cluster.
The Hidden Crisis in Sample Science
Physical samples are the unsung heroes of scientific research. From understanding climate change through ice cores to discovering new medicines from soil microbes, samples drive innovation across disciplines. Yet, we're facing a critical problem: most samples disappear into a "black hole" after collection.
Consider this: The University of Michigan Museum of Zoology manages over 150,000 specimens. When researchers tried to track how these specimens were used in publications, they found that only 245 out of 1,297 papers properly cited the specimens. The rest? Lost to poor documentation practices, making it impossible to understand the true impact of these invaluable collections.
Why This Matters for Open Science
The new ESIP guidelines address four critical challenges:
1. Enabling Large-Scale Sample Discovery
When researchers can efficiently cite hundreds or thousands of samples in their work, we can finally see the big picture of how samples contribute to science. The IEDA2 system already links over 34,000 unique samples to datasets—imagine this scaled globally!
2. Giving Credit Where It's Due
Collection managers, field researchers, and laboratories invest enormous resources in sample collection and curation. Proper tracking means they can finally demonstrate their impact to funders and institutions, ensuring continued support for these critical resources.
3. Tracking the Data Journey
A single sample might generate genomic data at one lab, chemical analyses at another, and ecological observations in the field. Current systems lose these connections, but with persistent identifiers (PIDs), we can follow a sample's complete scientific journey.
4. Breaking Down Disciplinary Silos
Environmental samples often cross disciplines—a soil sample might be relevant to microbiologists, geochemists, and climate scientists. Better tracking enables unexpected discoveries at these intersections.
The Power of Persistent Identifiers
The solution centers on PIDs—globally unique identifiers that work like DOIs for publications but for physical samples. Just as you can always find a paper using its DOI, PIDs ensure samples remain findable and citable forever. Over 12.5 million IGSN IDs have already been created, showing the momentum behind this movement.
What This Means for Researchers
The guidelines provide practical steps any researcher can implement today:
- Describe samples with rich metadata using community standards
- Assign or use existing PIDs for all samples
- Include PIDs in datasets and publications
- Cite samples properly in papers, making them discoverable
Building Tomorrow's Research Infrastructure
This isn't just about better bookkeeping—it's about transforming how we do science. When samples are properly tracked:
- Synthesis studies become possible across massive scales (think GBIF's 3.1 billion species records)
- Interdisciplinary collaborations flourish as researchers discover relevant samples from other fields
- Irreplaceable samples (from sites that no longer exist or can't be resampled) become accessible to global research
- Research reproducibility improves as others can access the exact samples used in studies
Join the Movement
The future of sample-based research is collaborative, transparent, and interconnected. Whether you're a researcher planning field work, a collection manager preserving specimens, or a data scientist building the next generation of research tools, you have a role to play.
By adopting these practices, we're not just organizing data—we're unlocking the full potential of every sample collected, ensuring that the effort invested in field work continues to generate discoveries for generations to come.
Ready to transform your sample management? Start by exploring the full guidelines and join the growing community building the future of open sample science.
The physical samples we collect today are the scientific treasures of tomorrow. Let's make sure their stories are never lost.
Research Citation
Damerow, J.E., Raia, N.H., Stanley, V. et al. Opening doors to physical sample tracking and attribution in Earth and environmental sciences. Sci Data 12, 1047 (2025). https://doi.org/10.1038/s41597-025-05295-z
